I had run ChromoPainter/fineSTRUCTURE for 715 South Asians using only about 90,000 SNPs. I thought it would be a useful exercise to use more SNPs, so I had to drop the Reich et al dataset. That left me with 615 individuals and 418,854 SNPs.
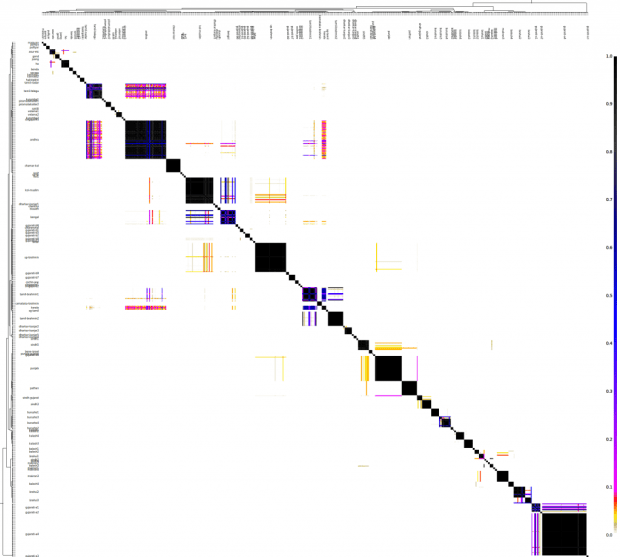
The "chunkcounts" file has the donors in columns and recipients in rows. Here's a heat map of the same.
fineSTRUCTURE classified these 615 individuals into 89 clusters. I have named these clusters for convenience, however, the names do not imply that anyone in the Punjab cluster is Punjabi.
While I created the cluster tree at the top of the spreadsheet, here's how the clusters are related.
The most interesting thing is how Gujarati A (likely Patels) are an out-group to everyone else. Another major grouping is that of the Baloch, Brahui and Makrani, along with 4 Sindhis (might be one of the Baloch tribe of Sindh?).
The Punjabis, Sindhis and Pathan get better classification here than they did last time.
The Punjab cluster includes 3 Gujarati B, 4 Pathans, 2 Singapore Indians, Punjabis, Haryanvis, Kashmiris, and a Rajasthani Brahmin. Even using this method, HRP0036, who is half-Sri Lankan and half-German/Polish was classified in the same cluster.
The Dharkar and Kanjar could not be separated at all here. According to Metspalu:
There are three second degree relatives groups in our sample: ..snip.. [Kanjar evo_37 and Dharkar HA023]. Again the last pair needs further explanation. The Dharkar and Kanjar practice a nomadic lifestyle and were living side by side at the time of sampling. As the ethnic border between the two is permeable we cannot rule out neither our error during sample collection and/or subsequent labelling nor shifted self-identity.
The inter-cluster heat map:
And you can see the chunkcounts donated from each cluster to recipient individuals in a spreadsheet.
The pairwise coincidence:
And the PCA plots:









































Recent Comments